|
|
R 1.-.-.- : Hydrolysis (bond cleavage by water) (Reaction)
L 1.13.-.- : Amide bond (One of Carbonyl Ester bonds; including peptide bond) (Ligand group involved)
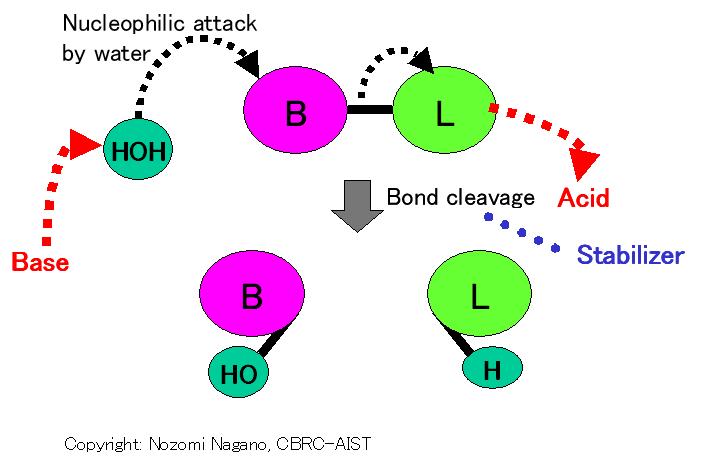
C 1.13.200.- : Pepsin-like mechanism (Acid-Base/Stabilizer); Water activation by base; SN2-like nucleophilic attack by activated water; Stabilization of transition-state; Protonation to leaving group by acid (Catalytic mechanism)
P 1.13.200.966 : Two acidic residues (interacting through a water) (Residues/cofactors in Protein)
1st Nucleophile : water
Catalytic groups : groups in residue/substrate
General Base : a catalytic residue
General Acid : a catalytic residue
Related Enzymes
There are 18 entries in this class.
D00231 : 3.4.23.25; Saccharopepsin (Catalytic domain; 2.40.70.10)
D00423 : 3.4.23.4; Chymosin (Catalytic domain; 2.40.70.10)
D00436 : 3.4.23.1; Pepsin A (Catalytic domain; 2.40.70.10)
D00437 : 3.4.23.3; Gastricsin (Catalytic domain; 2.40.70.10)
D00438 : 3.4.23.15; Renin-2 (Catalytic domain; 2.40.70.10)
D00439 : 3.4.23.20; Penicillopepsin (Catalytic domain; 2.40.70.10)
D00440 : 3.4.23.21; Rhizopuspepsin (Catalytic domain; 2.40.70.10)
D00441 : 3.4.23.22; Endothiapepsin (Catalytic domain; 2.40.70.10)
D00442 : 3.4.23.23; Mucorpepsin (Catalytic domain; 2.40.70.10)
D00443 : 3.4.23.24; Candidapepsin (Catalytic domain; 2.40.70.10)
D00444 : 3.4.23.39; Plasmepsin-2 (Catalytic domain; 2.40.70.10)
D00445 : 3.4.23.40; Phytepsin (Catalytic domain; 2.40.70.10)
D00471 : 2.7.7.7, 2.7.7.49, 3.1.26.4, 3.4.23.16; Gag-Pol polyprotein (Catalytic domain; 2.40.70.10)
D00484 : 3.4.23.5; Cathepsin D (Catalytic domain; 2.40.70.10)
D00462 : 3.5.1.52; Peptide-N(4)-(N-acetyl-beta-D-glucosaminyl)asparagine amidase F (Catalytic domain; 2.60.120.230)
D00529 : 3.4.23.-; Pepsin-2B (Catalytic domain; 2.40.70.10)
M00206 : (reaction 1) 2.7.7.49, 3.1.26.4, 3.4.23.-, 3.6.1.23; Pol polyprotein (Catalytic domains; 2.40.70.10, 2.70.40.10)
M00166 : (reaction 1) 2.7.7.7, 2.7.7.49, 3.1.26.4, 3.4.23.47; Gag-Pol polyprotein (Catalytic domains; 2.40.70.10, 3.30.70.270, 3.30.420.10)
|
|



