RLCP: Tutorial

Hierarchic Classification of Enzyme Catalytic Mechanisms

Description of the EzCatDB Database
The EzCatDB database analyzes and classifies the enzyme catalytic mechanisms on the basis of information from the literature and derived data from entries in the Protein Data Bank (PDB). Each data set contains corresponding enzyme information, such as E.C. number, PDB entries with their annotated ligand information and active site residues, information on catalytic mechanisms, and links to other databases, such as Swiss-prot, CATH, KEGG, Catalytic Site Atlas (CSA), MACiE, PDBsum, PDBj and PubMed.
Description of the RLCP classification
Hierarchic Classification of Catalytic Mechanisms, RLCP:
This is a part of the above EzCatDB database and a novel classification of enzyme catalytic mechanisms, which classifies the mechanisms at four levels:
R: Basic Reaction,
L: Ligand group involved in catalysis,
C: Catalysis type, and
P: Residues/cofactors located on Proteins.
"Basic reactions" are reaction types such as "hydrolysis", "phosphorolysis", and "transfer", which are mostly related to the primary number of Enzyme Commission (E.C.) numbers. At the second level (L), reactive parts of ligand molecules are considered. At the third level (C), the catalytic mechanisms are systematically classified, based on the information on existence of cofactors, types of nucleophile, acid, base, stabilizer, and modulator, SN2/SN1 (or associative/dissociative) reactions, and on how these catalytic groups function. The residues/cofactors on proteins involved in catalytic reactions are classified at the fourth level (P).
References
Nagano N., Nakayama N., Ikeda K., Fukuie M., Yokota K., Doi T., Kato T., Tomii K. (2015) "EzCatDB: the e
nzyme reaction database, 2015 update." Nucleic Acids Research, 43, D453-458.
Nagano N., Noguchi T., Akiyama Y. (2007) "Systematic Comparison of Catalytic Mechanisms of Hydrolysis and Transfer Reactions Classified in the EzCatDB Database."
PROTEINS: Structure, Function, and Bioinformatics. 66, 147-159.
Nozomi Nagano (2005) "EzCatDB" Nucleic Acids Research, Molecular Biology Database on-line compilation 2005, 6.1. Enzymes and Enzyme Nomenclature, 613.
Nozomi Nagano (2005) "EzCatDB: The Enzyme Catalytic-Mechanism Database."
Nucleic Acids Research, 33, Database Issue, D407-D412.
Differences between the RLCP classification and the conventional Enzyme Commission (E.C.) numbers
Whereas the conventional E.C. numbers are a kind of nomenclature of enzymes, the RLCP classifies the catalytic reactions, which considers the detailed catalytic mechanisms and protein structures as well as ligand information. Therefore, some enzymes might have several RLCP classes, rather than only one class, if they catalyze more than one reaction.
Furthermore, whole chemical structures of ligands (substrates and products) have been considered by the E.C. numbers, the RLCP has specifically addressed the reactive aspects of ligand molecules. Therefore, the first two levels of the RLCP classification (the R and L levels) can be correlated to the E.C. numbers.
Definition of "Basic Reaction" and "Ligand group involved in catalysis"
Hydrolysis:
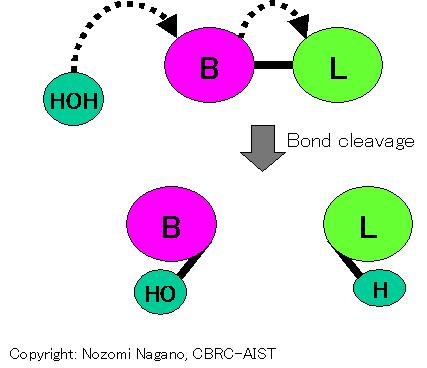
A reaction that cleaves a "bond" (B-L) with a water molecule. Ligand group involved in catalysis is the "bond".
 Phosphorolysis:
A reaction that cleaves a "bond" with a phosphate molecule. Ligand group involved in catalysis is the "bond".
Transfer:
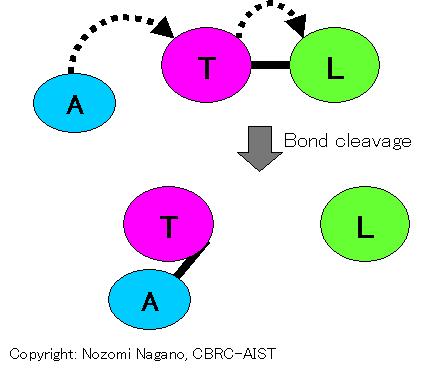
Reaction that transfers a "transferred group" (T) to an "acceptor group" (A) from a "leaving group" (L). The ligand group involved in catalysis is the "transferred group", the "acceptor group" and the "leaving group".
Phosphorolysis:
A reaction that cleaves a "bond" with a phosphate molecule. Ligand group involved in catalysis is the "bond".
Transfer:
Reaction that transfers a "transferred group" (T) to an "acceptor group" (A) from a "leaving group" (L). The ligand group involved in catalysis is the "transferred group", the "acceptor group" and the "leaving group".
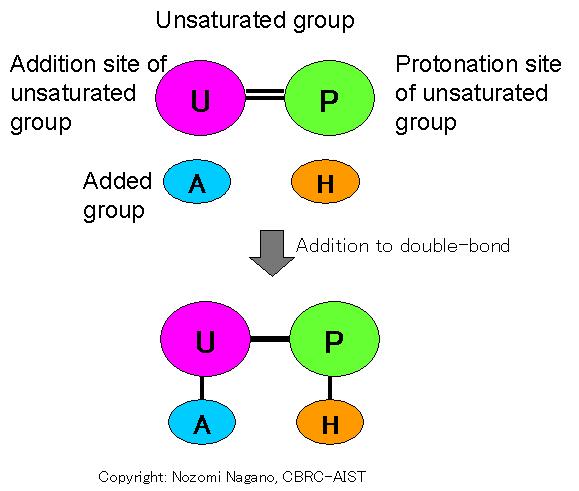
 Additive double-bond deformation: Reaction that adds an "added group" (A) to a "unsaturated group" with double-bond (U=P), in which the double-bond is converted to a single-bond. Ligand group involved in catalysis is the "Added group" and the "Unsaturated group". This reaction is inverse reaction of "eliminative double-bond formation", described below.
Additive double-bond deformation: Reaction that adds an "added group" (A) to a "unsaturated group" with double-bond (U=P), in which the double-bond is converted to a single-bond. Ligand group involved in catalysis is the "Added group" and the "Unsaturated group". This reaction is inverse reaction of "eliminative double-bond formation", described below.
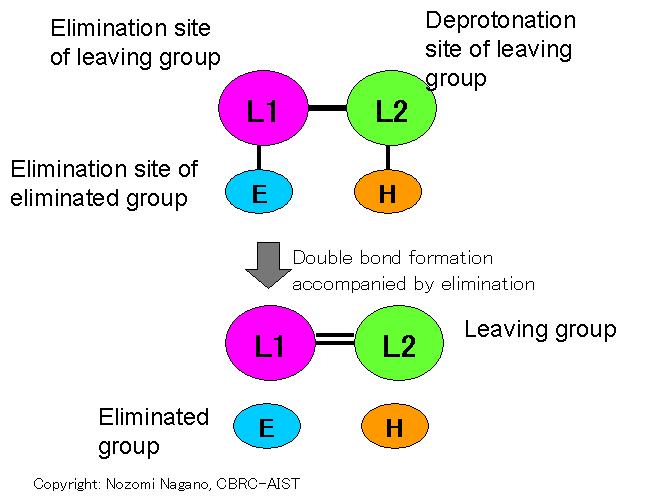
 Eliminative double-bond formation: Reaction that eliminates an "eliminated group" (E) from a "leaving group" (L1-L2), in which a double-bond is converted from a single-bond. Ligand group involved in the catalysis is the "eliminated group" and the "leaving group". This reaction is inverse reaction of "Additive double-bond deformation", described above.
Eliminative double-bond formation: Reaction that eliminates an "eliminated group" (E) from a "leaving group" (L1-L2), in which a double-bond is converted from a single-bond. Ligand group involved in the catalysis is the "eliminated group" and the "leaving group". This reaction is inverse reaction of "Additive double-bond deformation", described above.
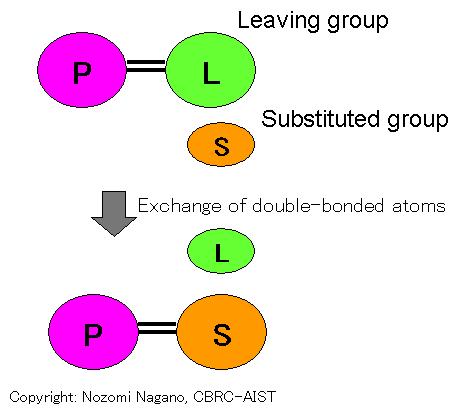
 Exchange of double-bonded atoms: Reaction that replaces double-bonded atoms. This reaction includes Schiff-base formation/deformation, which replaces carbonyl group by Schiff-base bond, and vice versa, and external/internal aldimine formation. Ligand group involved in catalysis is the "substituted group" (S) and the "leaving group" (L).
Exchange of double-bonded atoms: Reaction that replaces double-bonded atoms. This reaction includes Schiff-base formation/deformation, which replaces carbonyl group by Schiff-base bond, and vice versa, and external/internal aldimine formation. Ligand group involved in catalysis is the "substituted group" (S) and the "leaving group" (L).
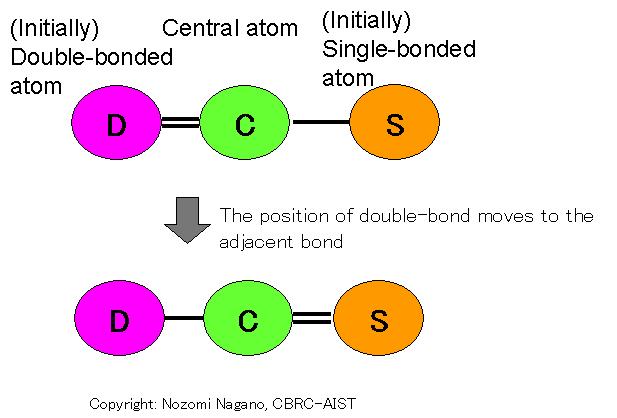
 Isomerization (shift of double-bond position): Reaction that changes the position of double-bond. Initial state and final state of ligand groups involved are annotaed at the "Ligand" level.
Isomerization (shift of double-bond position): Reaction that changes the position of double-bond. Initial state and final state of ligand groups involved are annotaed at the "Ligand" level.
 Hydride transfer: One of major redox reactions. This reaction catalyzes transfer of one proton and two electrons between substrate/intermediate and cofactor such as NAD(H) and NADP(H). This reaction is distinct from the electron transfer described below.
Electron transfer: One of major redox reactions that catalyzes transfer of electron, usually between metals or sulfur atoms that are involved in redox reactions.
Hydride transfer: One of major redox reactions. This reaction catalyzes transfer of one proton and two electrons between substrate/intermediate and cofactor such as NAD(H) and NADP(H). This reaction is distinct from the electron transfer described below.
Electron transfer: One of major redox reactions that catalyzes transfer of electron, usually between metals or sulfur atoms that are involved in redox reactions.
|
|
Copyright: Nozomi Nagano, JST & CBRC-AIST
E-mail to: Nozomi Nagano 
|