|
|
R 1.-.-.- : Hydrolysis (bond cleavage by water) (Reaction)
L 1.13.-.- : Amide bond (One of Carbonyl Ester bonds; including peptide bond) (Ligand group involved)
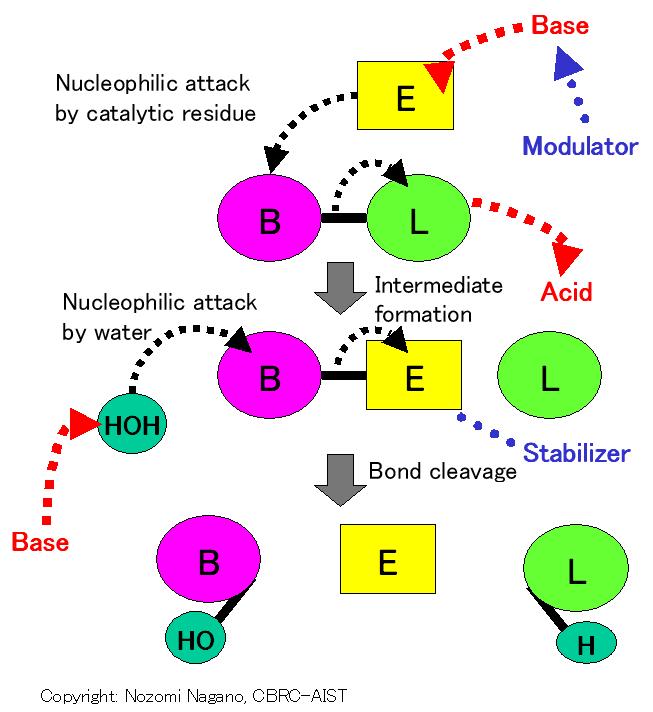
C 1.13.30000.- : Trypsin-like mechanism (Nucleophile/Acid-Base/Modulator/Stabilizer); Modulator-assisted base activation of nucleophile; Nucleophilic attack on target bond; SN2-like reaction; covalent bond between enzyme and substrate; followed by protonation to leaving group; hydrolysis by base-activated water (Catalytic mechanism)
P 1.13.30000.42 : Cys/His/Glu + mainchain amide (Residues/cofactors in Protein)
1st Nucleophile : a catalytic residue
Catalytic groups : groups in residue/substrate
General Base : a catalytic residue
General Acid : a catalytic residue
Related Enzymes
There are 2 entries in this class.
T00114 : (reaction 1) 6.3.5.2; GMP synthase {glutamine-hydrolyzing} (Catalytic domains; 3.40.50.620, 3.40.50.880)
M00217 : (reaction 2) 2.7.7.48, 3.4.22.28, 3.4.22.29, 3.6.1.15; Genome polyprotein (Catalytic domain; 2.40.10.10)
|
|



