| | How to use the  Please upload PDB-formatted protein structure data from your computer.
Please click on the "submit" button.
During the computation, the following display will appear. 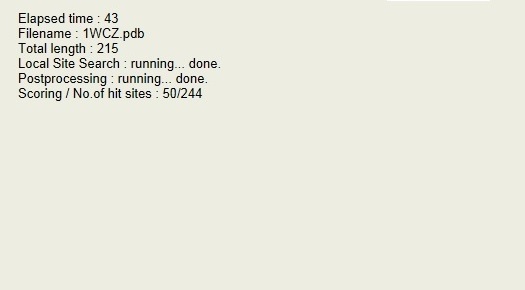
The result will be shown in the following way.
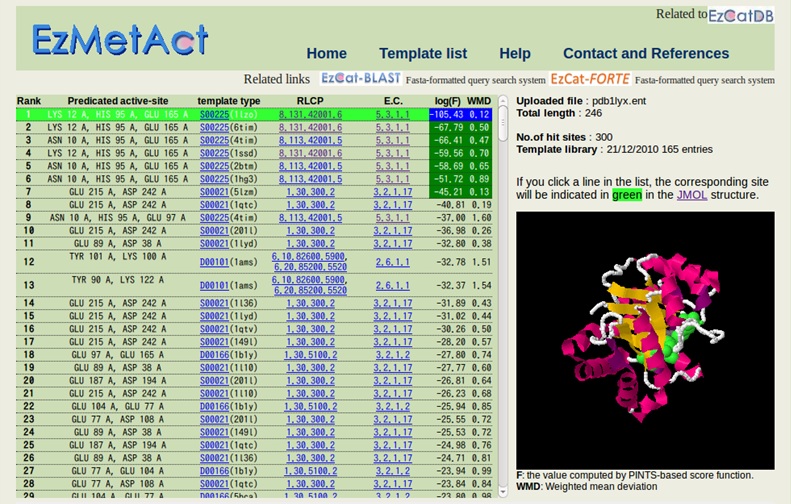
The rank 1 is selected and indicated in green. The active site is also indicated with spacefill model in green, along with the whole cartoon structure, on Jmol at the right hand.
The significant hits are indicated with the colored scores (log(F) and WMD). In this figure, the ranks up to the seventh are significant. If you select rank 9 by clicking on it, the rank 9 will be indicated in green, along with the corresponding structure on Jmol.
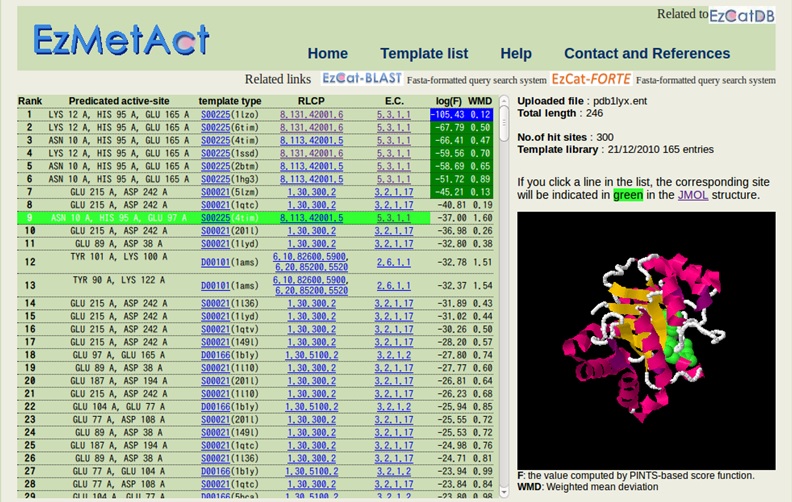
For the Jmol viewer, please install java in your machine, since the software is a Java applet.
For Windows machines, you will have to enable the Java contents at Java Control Panel.
In order to enable the Java contents, please select "Configure Java" from "All Programs" of "Start Menu",to open the Java Control Panel. Please check the checkbox for "Enable Java content in the browser", make the security level a "Medium" level, and click on the "Apply" button at the Java Control Panel.
The Java documentation by Oracle will be very useful for the java setup.
|