Basic information
This sequence search system can be used to search against EzCatDB database (Nagano, 2005), using FORTE algorithm (Tomii & Akiyama, 2004). This system is suitable for detecting remote homologues.
Please cite the following references.
References:
Nagano, N., Nakayama, N., Ikeda, K., Fukuie, M., Yokota, K., Doi, T., Kato, T., Tomii, K. "EzCatDB: the enzyme reaction database, 2015 update." Nucleic Acids Research, in press.
Tomii, K. & Akiyama, Y. (2004) "FORTE: a profile-profile comparison tool for protein fold recognition." Bioinformatics, 20, 594-595. PubMed link
Nagano, N. (2005) "EzCatDB: The Enzyme Catalytic-Mechanism Database." Nucleic Acids Research, 33, D407-D412.
Tomii, K., Hirokawa, T., and Motono, C. (2005) "Protein structure prediction using a variety of profile libraries and 3D verification." Proteins, 61, 114-121. PubMed link
How to use EzCat-FORTE
If you submit your fasta-formatted query and your e-mail address to this system, you will receive message from webmaster@aist.go.jp that your EzCat-FORTE job is finished. In the message, your query password, which can be searched in the "Show Results" page, is described, along with the direct links to your result. The result will be stored up to one week.
It usually takes around ten minutes for a sequence with around 300 residues, unless this server is too busy.
Although the sequences with up to 2000 residues can be calculated in terms of the FORTE algorithm, those with up to 800 residues are recommended as query sequences, due to the capacity of this server.
The hits with the Z-scores higher than 7.0 are desirable ones in this system, although you can see the hits with the lower Z-scores.
An example of the result page:
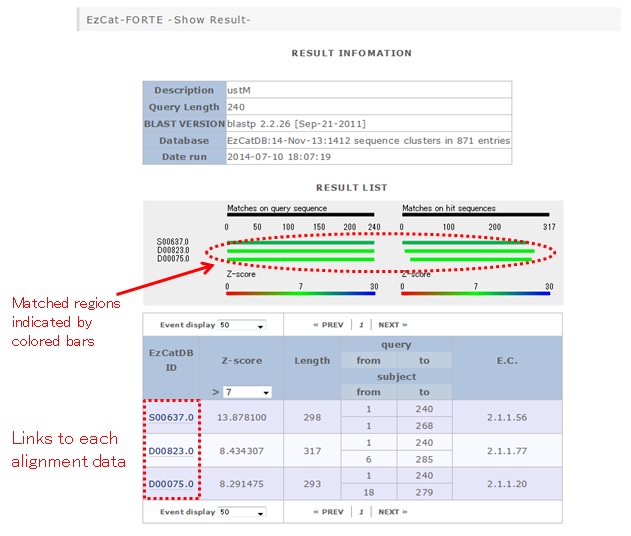
If you click the link to the alignment data, the following page will appear:
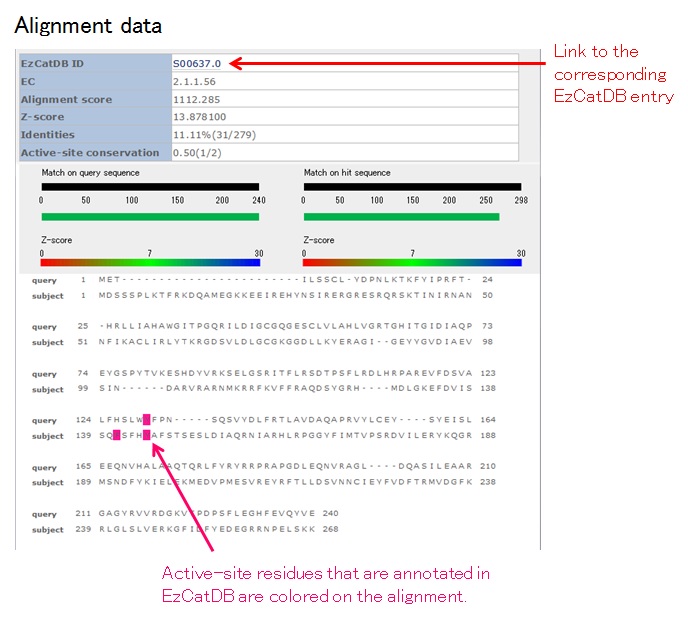
In the alignment data, functional residues in EzCatDB are colored as follows:
Catalytic residues (sidechain): magenta
Catalytic residues (mainchain): green
Cofactor-binding residues: orange
Modified residues: yellow
Regarding "Active-site conservation" in Alignment data, currently, the number of the same matched residues is simply divided by the total number of active-site residues in the hit-representative sequences, although we are planning to consider the chemical similarities between the matched residues in the future.
New function: Comparison with the results by EzCat-BLAST (April 2016 ~)
The new system provides the comparison with the results by EzCat-BLAST. The hit sequence date can be accompanied by the following symbols:
 The hit sequence is detected also by EzCat-BLAST, and its rank in EzCat-FORTE is higher than that in EzCat-BLAST.
The hit sequence is detected also by EzCat-BLAST, and its rank in EzCat-FORTE is higher than that in EzCat-BLAST.
 The hit sequence is detected also by EzCat-BLAST, and its rank in EzCat-FORTE is lower than that in EzCat-BLAST.
The hit sequence is detected also by EzCat-BLAST, and its rank in EzCat-FORTE is lower than that in EzCat-BLAST.
 The hit sequence is detected also by EzCat-BLAST, and its rank in EzCat-FORTE is the same as that in EzCat-BLAST.
The hit sequence is detected also by EzCat-BLAST, and its rank in EzCat-FORTE is the same as that in EzCat-BLAST.
 The hit sequence is detected only by EzCat-FORTE, not by EzCat-BLAST.
The hit sequence is detected only by EzCat-FORTE, not by EzCat-BLAST.