|
|
How to use the

Please input FASTA-formatted sequence data in the following way.

Please click on the "submit" button.
(When the radio button next to "Swiss-Prot with Active site/Binding site or with EC numbers" is selected, the BLAST search against the Swiss-Prot database will start [See below].)
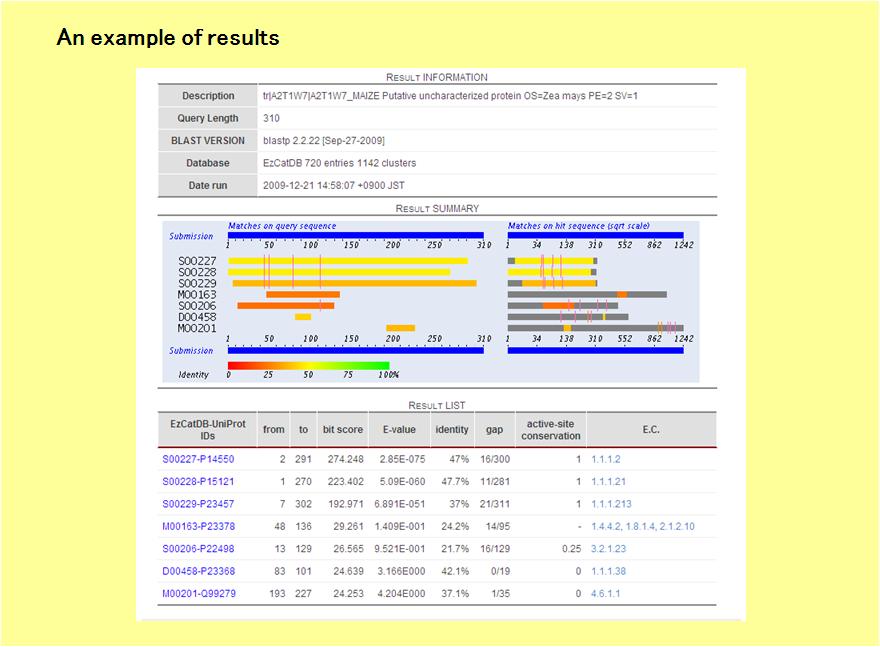
The result will be shown in the following way.
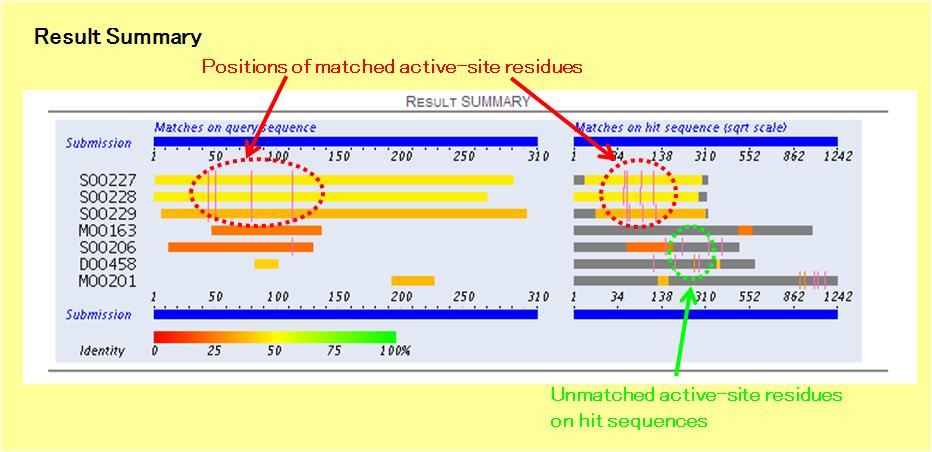
In the Result Summary, the positions of active-site residues, which are annotated for the hit sequences from EzCatDB, are shown as colored lines. The unmatched active-site residues are shown on the gray regions on the right-hand diagram, but the matched active-site residues are on the colored regions, which are the matched sequence regions, on both diagrams.
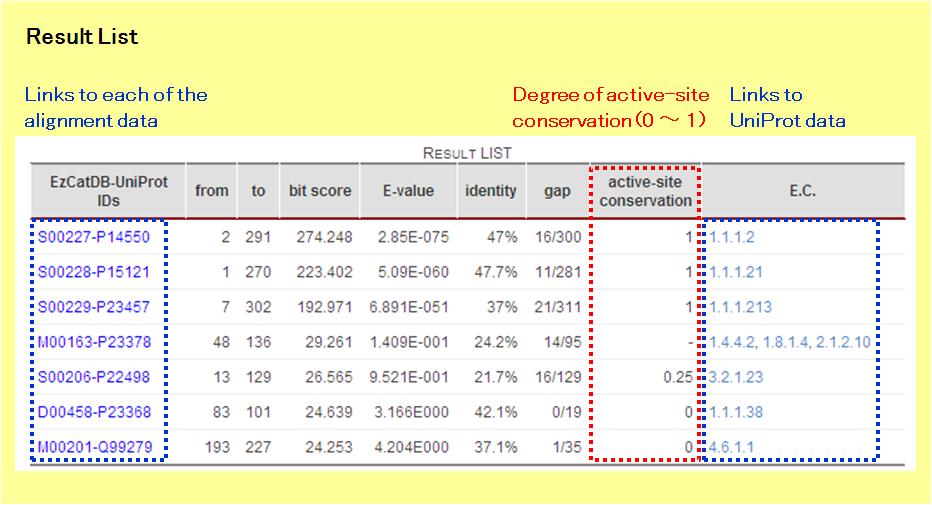
In the Result List, the information related to the hit sequences is presented. In the EzCat-BLAST, the active-site conservation is considered. If the hit sequence includes any annotated active-site residues in the EzCatDB entry, then the active-site conservation must be from 0 to 1. If the figure is one, then the active-site residues are conserved completely. On the other hand, if the hit sequence contains no active-site residue, then the active-site conservation will not be calculated.
There are links to the alignment data along with those to the NiceZyme data from UniProt. By clicking the corresponding "EzCatDB-UniProt" IDs, the alignment data will open.
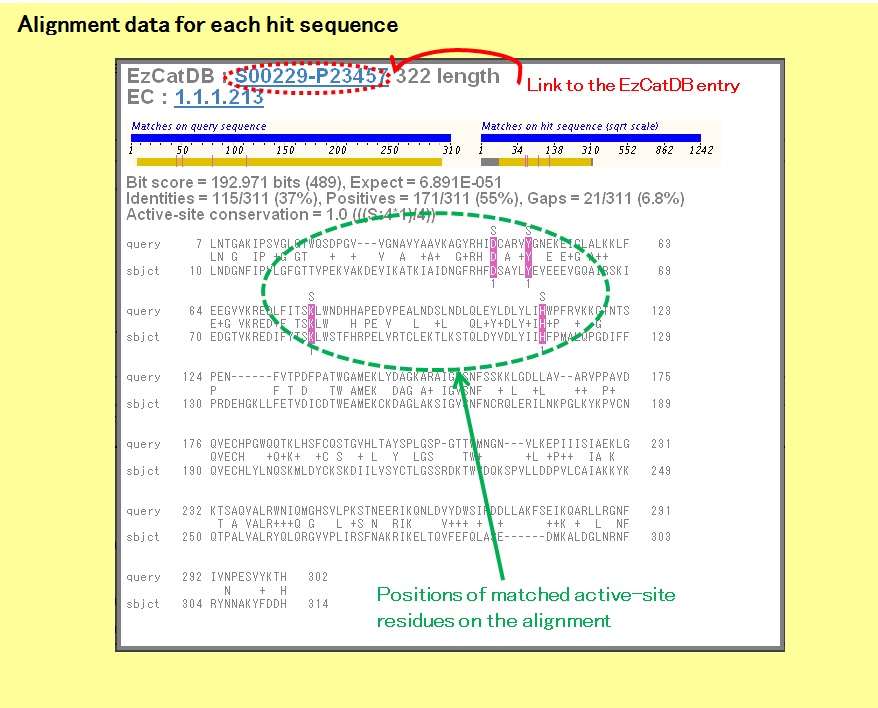
In the alignment data, the matched active-site residues are colored. There is also a link to the corresponding EzCatDB entry.
By pointing the cursor to the active-site residues, detailed information related to the active-site residues will be shown.
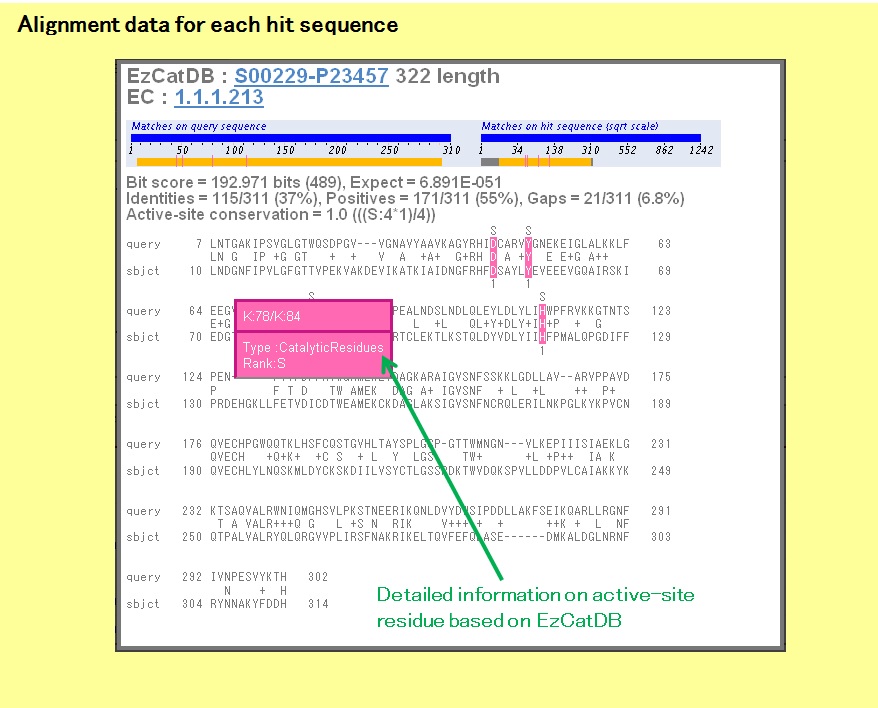
Here, if the matched residue on the hit sequence is the same as that of the query sequence, then the S rank will be assigned to the matched residue. If the matched residue on the hit sequence is chemically similar to that of the query sequence, then the A rank will be assigned to the matched residue. Otherwise, the B rank will be assigned to the matched residue. These ranks will be considered in the active-site conservation.
Functional residues in EzCatDB are colored as follows:
Catalytic residues (sidechain): magenta
Catalytic residues (mainchain): green
Cofactor-binding residues: orange
Modified residues: yellow
A similar analysis can be applied to the Swiss-Prot sequence database with functional residues such as Active site, Metal binding site, Binding site, and Modified Residue, or with EC numbers. For performance, up to 30 sequences can be shown for the Swiss-Prot analysis.
Functional residues in Swiss-Prot are colored as follows:
Active site: magenta
Metal binding site or Binding site: orange
Modified residues: yellow
System developed & maintained by
Naofumi Sakaya (IMS Lab.Inc.)
|